1. Introduction
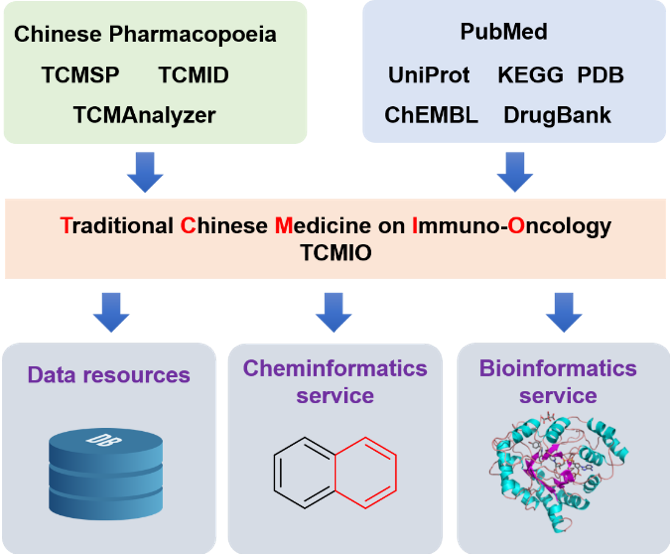
Previous studies have illustrated the important roles of TCMs in immune regulation and proposes a promising future for them in cancer immuno-therapies. In this work, we collected the IO targets and their small molecule ligands, those ligands information were further mapped with the chemical ingredients of the TCM. All those collected data were deposited in web-based publicly accessible database TCMIO, cheminformatics and bioinformatics tools were integrated into this database for user customer analysis.
2. Architecture of TCMIO
TCMIO was implemented into a web-based publicly accessible database, which can be accessed through major web browser. Beego, an open source framework for rapid web development in Golang, used as backend services. RESTful APIs are provided in TCMIO. All data were stored in open source relational database PostgreSQL (version 10.5). The ChemDoodle web component is used for chemical structure visualization. Bingo is used as molecular search cartridge, owning to the state-of-the-art indexing algorithms within the underlying database server, and its supports for high performance similarity, substructure, and full structure searches functions. In order to investigate the mechanism of actions of TCM, Gene enrichment analysis tools were integrated into TCMIO. The Database for Annotation, Visualization and Integrated Discovery (DAVID) is integrated into TCMIO.
3. Tools used in TCMIO
| Tools | Purpose | Link |
|---|---|---|
| Chinese Pharmacopoeia(2015) | Pharmacopoeia(2015) prescriptions and TCM data source | wp.chp.org.cn/front/chpint/en |
| UniProt | IO taregts information | www.uniprot.org |
| ChEMBL 24 | IO targets and ligands | www.ebi.ac.uk/chembl |
| KEGG | pathways data source | www.kegg.jp |
| TCMAnalyzer | ingredients of TCM | www.rcdd.org.cn/tcmanalyzer |
| TCMSP | ingredients of TCM | lsp.nwsuaf.edu.cn/tcmsp.php |
| TCMID | ingredients of TCM | www.megabionet.org/tcmid |
| DAVID | go enrichment analysis | david.ncifcrf.gov |
| ipmDraw | structure draw | ipmdraw.iprexmed.com |
| ChemDoodle Web | structure display of natural products | web.chemdoodle.com |
| Vis.js | network display | visjs.org |
| PostgreSQL | storage database | www.postgresql.org |
| Golang | web server language | golang.org |
| JQuery | foreground and background interaction | jquery.com |
4. Functions
TCMIO contains three functional parts, Browse, Structure, and MOA. Browse page provides information on prescription, TCM, ingredient, target, and ligand libraries. Interactive tables with sorting and searching functions are provided for user to access the data conveniently. Structure page provides cheminformatics tools to accessed the ingredient and ligand libraries which contains the chemical structure data. MOA page provides the bioinformatics tools, concluding network and pathway analysis, to elucidating the mechanism of actions of TCM or prescription.
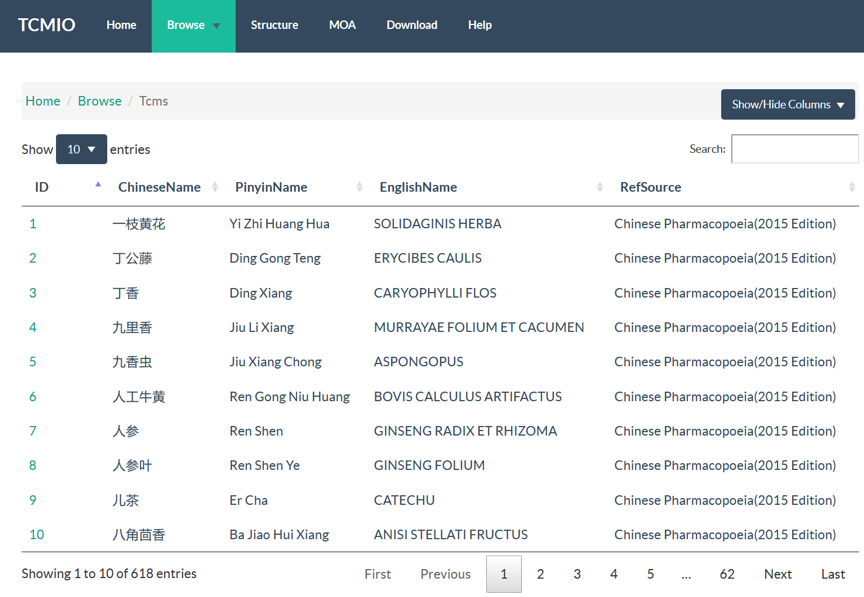
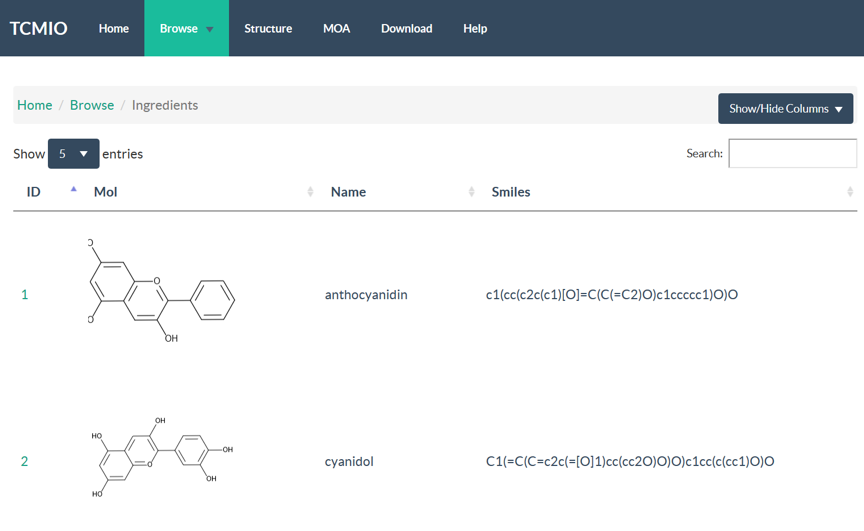
4.1 Browse
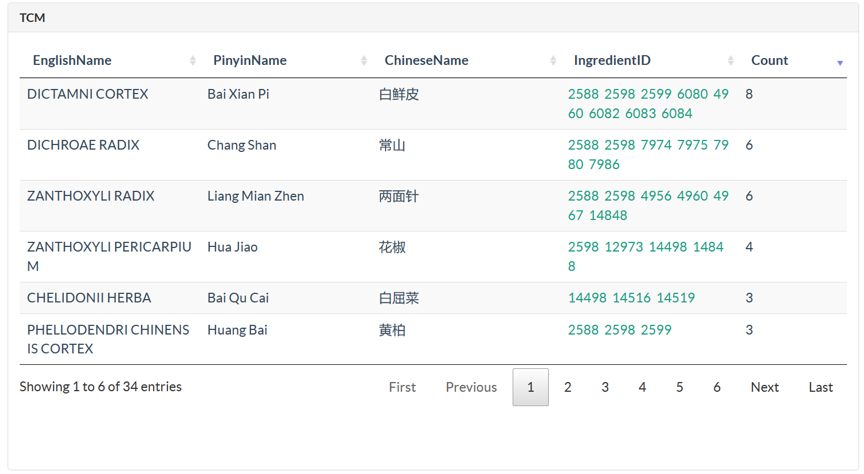
TCMIO contains prescription, TCM, ingredient, target, and ligand libraries. In Browse page, user can browse all the data entries with an interactive table. Server-side processing in DataTables is used, which provides powerful browsing, searching and sorting ability, which help user to access the data of interest efficiently. Searching function supports multi-columns. When user input the search words, server would search multiple columns of current library, and return the results with paging.
4.2 Structure
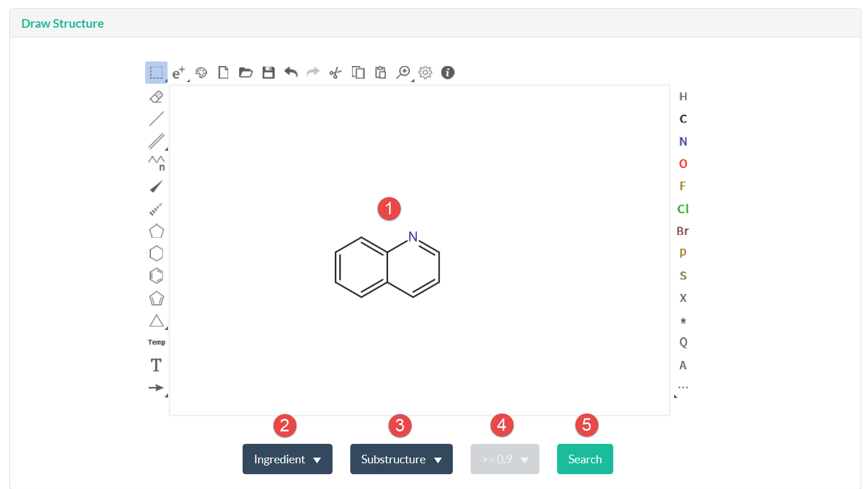
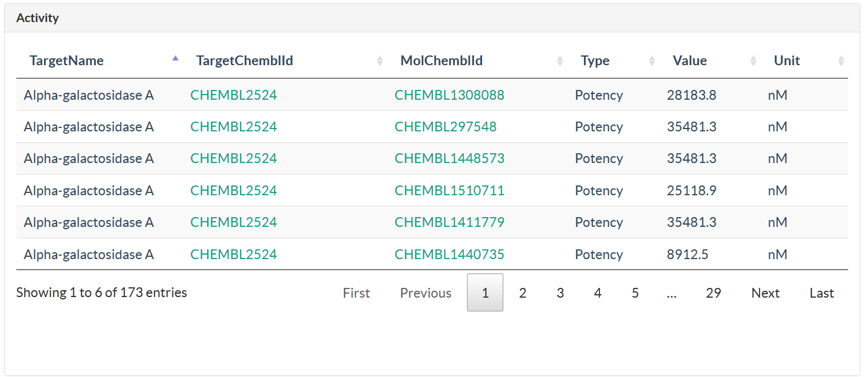
In Structure page, user could search against ligand and ingredient library using structure search methods, which contains full-structure, sub-structure, and similarity search. There are 5 steps for user to submit a structure search. Step 1: Draw a query structure, or upload structure file using ipmDraw. Step 2: Select a library, ingredient or ligand. Step 3: Select a structure search method. Step 4: Select a similarity threshold. This is only for similarity search. Step 5: Click Search button. Structure search against ingredient library, user could identify all the TCMs which contains the specific chemical structure or similar ingredients with the query structure. Structure search against ligand library, user could identify all ligand hits and their targets with bioactivity data.
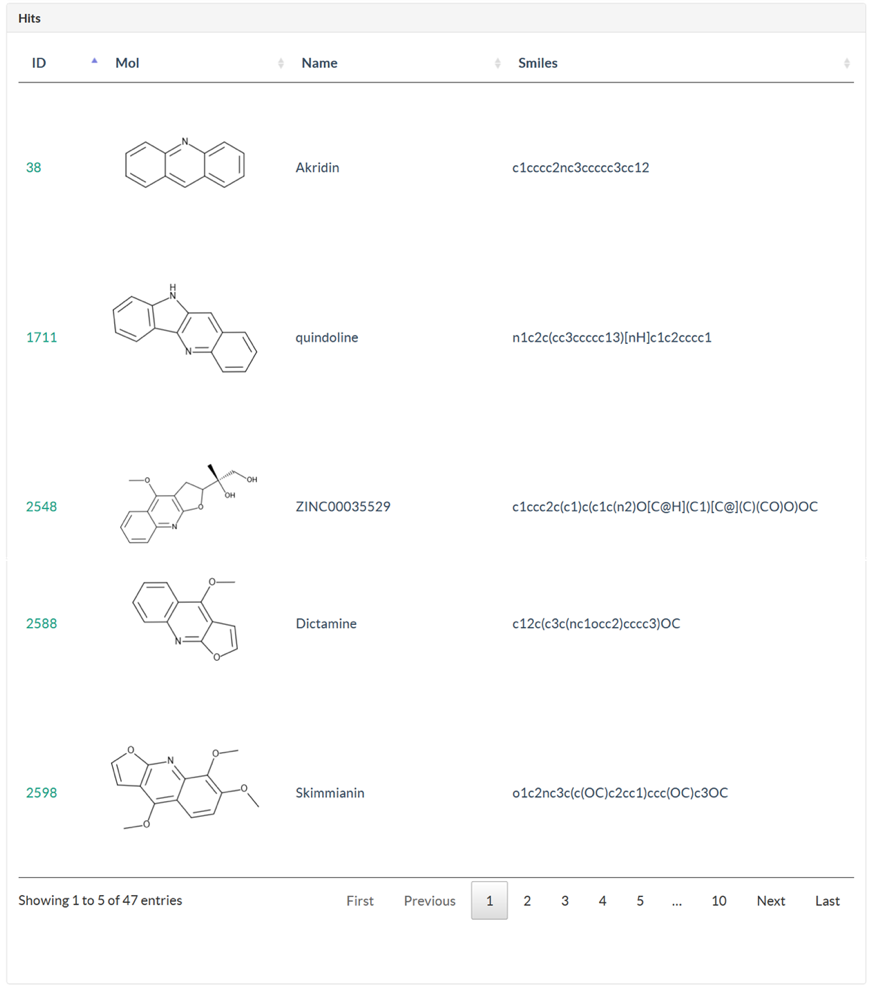
4.3 MOA
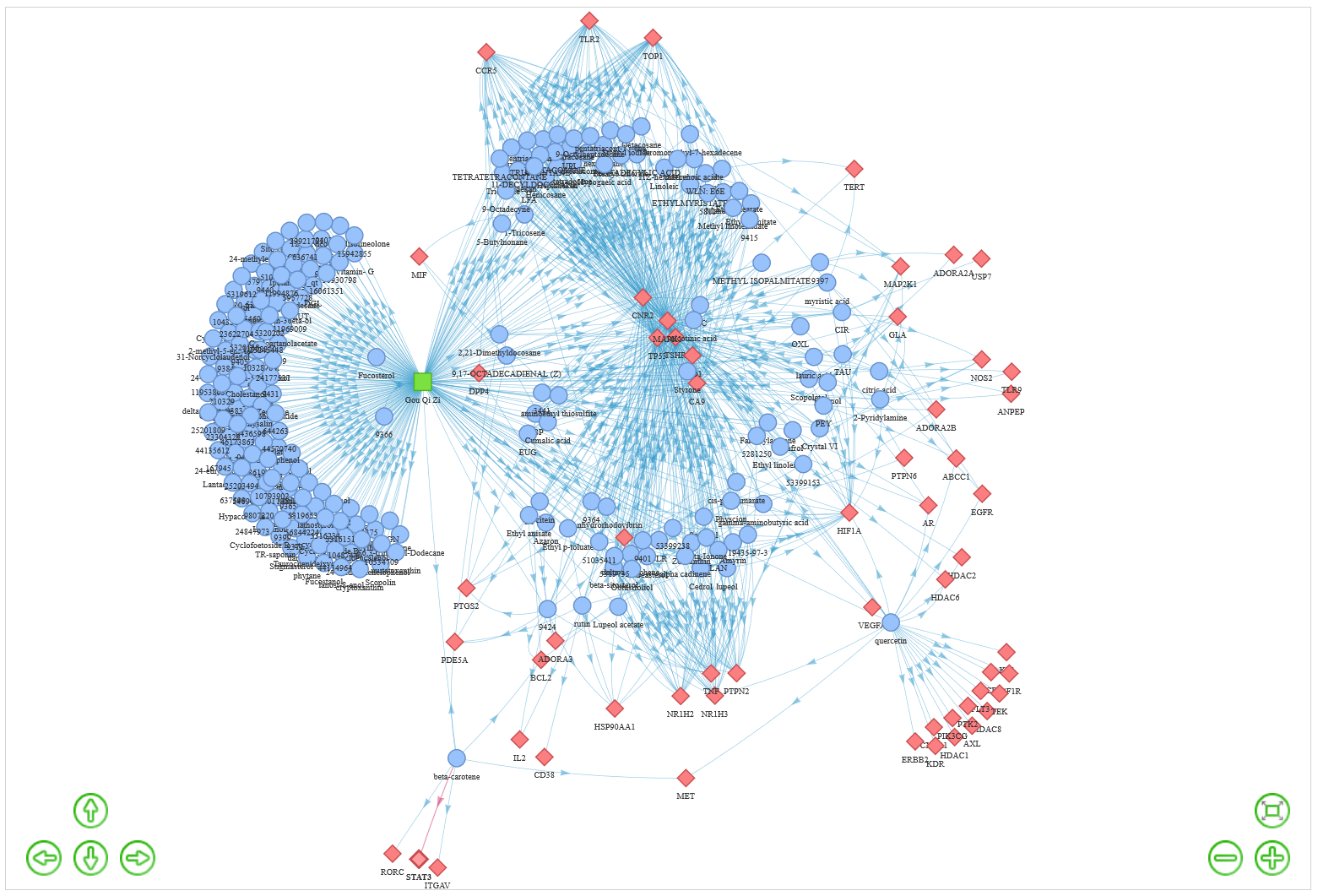
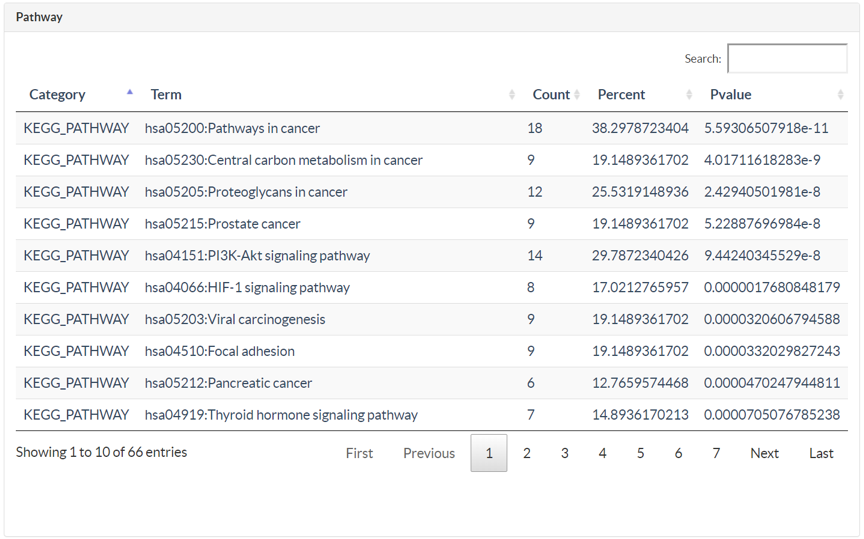
In MOA page, user could submit TCMs or Prescription with different name types. TCMIO would construct a TCM-ingredient-target network automatically. User could identify the high degree ingredients and targets in the network. The high degree for ingredient suggesting the ingredient could interact with multiple targets, and the high degree for target indicating the target could be targeted by multiple ingredients. This network could conveniently elucidate the mechanism of action for TCM or prescription. Moreover, those targets could be subsequently submitted the DAVID web server for pathway enrichment analysis.
5. Download
All data in TCMIO could be downloaded from Download page.
About us
TCMIO was corporately developed by Gut Microbiome and Health (GMH) Group at Guangdong Institute of Microbiology, and Traditional Chinese Medicine and Network Medicine (TCMNM) Group at Guangzhou University of Chinese Medicine.
GMH was established by Dr. Liwei Xie. Dr. Xie was trained in the area of both Molecular Nutrition and Computational Biology in United States in 2007-2017. The research group focuses on the AI-algorithm development and application in drug design, functional and health benefit small molecules and nature products design and screening, targeting gut microbial dysbiosis-associated metabolic diseases, such as Type 2 diabetes (T2D) and obesity.
TCMNM was established by Dr. Jiansong Fang. Dr. Fang has a postdoctoral training (funded by China Scholarship Council) on network medicine at Cleveland Clinic in United States. His group combines tools from genomics, network medicine, systems pharmacology, chemical biology and experimental pharmacology to address the challenging questions of deciphering the mechanisms of TCM or their ingredients against various human complex diseases (e.g., Cancer and Alzheimer’s disease).
Address
Dr. Liwei Xie, xielw@gdim.cnState Key Laboratory of Applied Microbiology Southern China
Guangdong Institute of Microbiology
100 Xianlie Middle Rd, BLD58 2rd Floor
Guangzhou, Guangdong, China, 510070
Dr. Jiansong Fang, fangjs@gzucm.edu.cn
Science and Technology Innovation Center
Guangzhou University of Chinese Medicine
12 Jichang Rd, Baiyun District
Guangzhou, Guangdong, China